SAAM II
SAAM II (an abbreviation of "Simulation Analysis and Modeling" version 2.0) is a proprietary and US NIH-funded software that was developed by SAAM Institute in 1997 at the University of Washington in Seattle. SAAM II is a software for tracer and pharmacokinetic studies but also deals easily with all types of compartmental and noncompartmental models in order to help researchers create models, design and simulate experiments, and analyze data quickly, easily, and accurately. Compartmental models are constructed graphically using drag and drop icons. The models can be either linear or nonlinear (kinetics). From the easy design of a model and instructions to run the model ("experiment"), SAAM II automatically creates a system of ordinary differential equations which are solved using powerful stiff and non-stiff solvers.
 | |
| Developer(s) | Nanomath LLC |
|---|---|
| Initial release | v1.0 1995 |
| Stable release | v2.3 2019
|
| Written in | C++ |
| Operating system | Windows, macOS |
| Platform | IA-32, x86-64 |
| Type | Compartmental modeling |
| License | Proprietary commercial software |
| Website | www |
SAAM II can handle complex problems and has no built-in upper limit on the size of the model. It can handle multiple, simultaneous data sets and experiments. This makes it unique in the sense that other PK software, like WinNonLin, are usually set for the typical 1 or 2 compartment problems. It was developed by scientists and engineers whose main expertise is in the modeling of lipids or glucose, but the same functionality is required for PK and PK/PD modeling. Hence, it is considered in the field of pharmacokinetics, nutrition, and radiolabel tracing. It is considered the easiest way to prototype models and analyze data. It was thought of as software to help nonmodelers simulate, fitting, and analyze their data. It counts thousands of users worldwide. It was validated in many fields, especially in the glucose kinetics for the artificial pancreas prototyping. It is also used in many routine [pharmacokinetics] assessments and its community is niche and very dear to this software.
History
University of Washington working through its Resource Center for Kinetic Analysis in the Center for Bioengineering, rewrote code for a software package called SAAM (I or 1.0), which had been developed in the late 1950s and early 1960s at the National Institutes of Health and at other research sites in the United States. The original SAAM was a powerful research tool to aid in the design of experiments and the analysis of data. Its dictionary permitted compartmental models to be developed without formally specifying the differential equations. However, SAAM was developed before the era of modem computer software design and implementation techniques, and as a result, it was not user-friendly. In addition, the computational kernel was not well documented, which made validation, maintenance, and further enhancements difficult.[1] In 1995 Prof David Foster led the re-writing of the SAAM code, including a strategic user interface, which generated SAAM II. It was then licensed for-profit concern. The SAAM Institute held the license until it was terminated in 2005. A particular boost to SAAM II was then given by both Prof Foster and Dr. Vicini's labs furthering the development and support, plus the creation of a Population pharmacokinetics plug-in, called PopKinetics. From 2011 to 2021 the distribution was licensed to a software company in Virginia, the Epsilon Group. They have updated SAAM to the current version 2.3, with bugs fixing, graphic improvements, and the creation of the Mac version.
Now, SAAM II is distributed by Nanomath LLC, a Washington company held by Dr. Simone Perazzolo a modeler and scientist at University of Washington Pharmaceutics.
Features
It has a user-friendly graphical user interface wherein compartmental models are constructed by creating a visual representation of the model. From this model, the program automatically creates systems of ordinary differential equations (ODEs). The program can both simulate and fit models to data, returning optimal parameter estimates and associated statistics. It is composed of a compartmental and numerical module.
The Compartmental Module
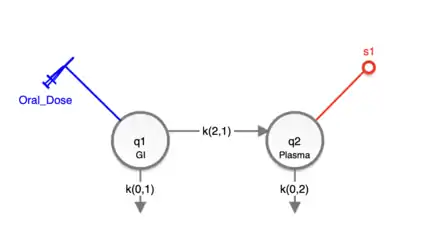
In the compartmental module, the user can choose from a set of drag and drop model-building icons representing compartments, transfers (0th-order, linear, Michaelis–Menten kinetics, etc.), and delays to build a visual representation of a system on a drawing canvas. For each icon, attributes can be defined using dialogue boxes. For example, reference names can be assigned to compartments. To simulate the behavior of the model, an experiment must be created (model conditions). Using the drag and drop experiment-building icons, the user directly specifies exogenous inputs and sample sites. Each has an attribute box where, for example, measurement equations are written and associations with data elements are made. The example of a two compartmental model for the oral absorption of a candidate drug (#1) and the plasma elimination (#2) is depicted in Figure 1. Note that the input (dose) is in the digestive compartment 1, while the output (observations) is made in the plasma compartment 2. The Compartmental Module is the most used feature of SAAM II.
The Non-compartmental Module (Numerical Module)
The Numerical module is also available but less frequently used; it lets you write directly the equations of the model or model directly the data by predefined functions. The latter allows you to carry out a non-compartmental analysis of the data. It requires coding.
Main Computational Algorithms
For ODE solving it uses 3 types of integrators: RK 4-5th order, something like ode45 in MATLAB; a method based on the Pade approximation of the matrix exponential; and a typical Rosenbrock method.
An appreciated feature of SAAM is the creation of a weighting scheme based on the supposed measurement error as well as the Bayesian fitting.[2] When selected, the Bayesian fitting is particularly powerful in parameter estimation accuracy. SAAM allows the estimation of complicated structures even where theoretical structural identification is not successful or too complex to assess. However, when the model tends to hard unidentifiability, SAAM pops a message as the precision of the estimates is not computable. This is a commonsense and practical feature of SAAM which gives an important leeway in parameter fitting of rather complex structures with few data observations, whereas other fitting software fails.
Validation
A validation of the software (numerical) performance was carried out against the industry gold-standard PK analysis software, Phoenix WinNonlin. In general, there was good agreement (<1% difference) between SAAM II and WinNonlin in terms of parameter estimates and model predictions[3]
popKinetics PlugIn
popKinetics is an add-on to SAAM II and a powerful simulator to simulate the PK of a drug in populations with variability in data and model parameters. It was funded by the NIH for the population analysis of the compartmental models built in SAAM II. It has a user interface that is easy to use (no programming or pseudo-code is needed). It computes the Standard Two-Stage and Iterated Two-Stage approaches for population parameters with their confidence intervals. Few if any assumptions about the population are necessary prior to analysis. For example, clinical trials can be simulated to determine the effect of varying dosing regimens.
Impact and Notable Work
The Type 1 Diabetes Simulator (AKA, Padua-Virginia Simulator) allows the simulation of the glucose and insulin profile of a type 1 diabetic.[4] In 2008 the US Food and Drug Administration accepted the T1D simulator developed by University of Virginia and University of Padova as a substitute for preclinical trials for certain insulin treatments, including closed-loop algorithms for artificial pancreas. Cobelli and Kovatchev labs prototyped the simulator in SAAM II. A SAAM II version of the simulator is avialble for charge.
Classic pharmacokinetic studies using SAAM II are by Foster and Vicini labs, while more complex lipid dynamics studies are by Barrett lab in Australia.[5] SAAM II is currently employed for prototyping PBPK models. When complex mechanisms participate in the PK of new drugs or formulations, SAAM II can be used for rapid in vivo testing. Perazzolo and colleagues in Seattle use SAAM II jointly with experimental data to inform nanoparticle PBPK and the clinical transition of long-acting or extended-release therapies.[6][7]
Education
SAAM II was present in several curricula in Universities in the US and around the world, especially in the early 2000. The University of Washington is still teaching SAAM II in the PK module at the School of Pharmacy. At the University of Padova, SAAM II is introduced to biomedical engineers in their graduate modeling modules.
References
- Cobelli, Claudio; Foster, David (1998). "Compartmental models: theory and practice using the SAAM II software system". Adv Exp Med Biol. Advances in Experimental Medicine and Biology. 445 (445): 79–101. doi:10.1007/978-1-4899-1959-5_5. ISBN 978-1-4899-1961-8. PMID 9781383.
- Barrett, P. Hugh R.; Bell, Bradley M.; Cobelli, Claudio; Golde, Hellmut; Vicini, Paolo; Foster, David M (1998). "SAAM II: Simulation, Analysis, and Modeling Software for Tracer and Pharmacokinetic Studies". Metabolism. 47 (4): 484–492. doi:10.1016/s0026-0495(98)90064-6.
- Heatherington, Anne C; Vicini, Paolo; Golde, Hellmut (1998). "A Pharmacokinetic/Pharmacodynamic Comparison of SAAM II and PC/WinNonlin Modeling Software". J Pharm Sci Biol. 87 (10): 1255–1263. doi:10.1021/js9603562. PMID 9758686.
- Dall Man, Chiara; Micheletto, Francesco; Lv, Dayu; Brenton, Marc; Kovatchev, Boris; Cobelli, Claudio (2014). "The UVA/PADOVA Type 1 Diabetes Simulator". J Diabetes Sci Technol. 8 (1): 26–34. doi:10.1177/1932296813514502. PMC 4454102. PMID 24876534.
- Barrett, P Hough R; Bell, Bradley; Cobelli, Claudio; Golde, Hellmut; Shumitzky, Alan; Vicini, Paolo (1998). "SAAM II: Simulation, analysis, and modeling software for tracer and pharmacokinetic studies". Metabolism. 4 (4): 484–492. doi:10.1016/S0026-0495(98)90064-6.
- Perazzolo, Simone; Sheriman, Laura; McConnachie, Lisa; Shen, Danny; Ho, Rodney J (2020). "Integration of Computational and Experimental Approaches to Elucidate Mechanisms of First-Pass Lymphatic Drug Sequestration and Long-Acting Pharmacokinetics of the Injectable Triple-HIV Drug Combination TLC-ART 101". J Pharm Sci. 109 (5): 1789–1801. doi:10.1016/j.xphs.2020.01.016.
- Perazzolo, Simone; Shen, Danny; Ho, Rodney J (2022). "Physiologically Based Pharmacokinetic Modeling of 3 HIV Drugs in Combination and the Role of Lymphatic System after Subcutaneous Dosing. Part 2: Model for the Drug-combination Nanoparticles". J Pharm Sci. 111 (3): 825–837. doi:10.1016/j.xphs.2021.10.009.